

Reconstructing a phylogenetic network for a set of conflicting phylogenetic trees on the same set of leaves remains an active strand of research in mathematical and computational phylogenetic since 2005, when Baroni et al. [1] showed that the minimum number of reticulations h(T,T') needed to simultaneously embed two rooted binary phylogenetic trees T and T' into a rooted binary phylogenetic network is one less than the size of a maximum acyclic agreement forest for T and T'. In the same paper, the authors showed that h(T,T') is bounded from above by n-2, where n is the number of leaves of T and T' and that this bound is sharp. That is, for a fixed n, there exist two rooted binary phylogenetic trees T and T' such that h(T,T')=n-2.
Since 2005, many papers have been published that develop exact algorithms and heuristics to solve the above NP-hard minimisation problem in practice, which is often referred to as Minimum Hybridisation in the literature, and that further investigate the mathematical underpinnings of Minimum Hybridisation and related problems. However, many such studies are restricted to two trees and much less is known about Minimum Hybridisation for when the input consists of more than two phylogenetic trees, which is the more relevant cases from a biological point of view.
In [2], van Iersel, Jones, and Weller establish the first lower bound for the minimum reticulation number for more than two rooted binary phylogenetic trees, with a focus on exactly three trees. The above-mentioned connection between the minimum number of reticulations and maximum acyclic agreement forests does not extend to three (or more) trees. Instead, to establish their result, the authors use multi-labelled trees as an intermediate structure between phylogenetic trees and phylogenetic networks to show that, for each ε>0, there exist three caterpillar trees on n leaves such that any phylogenetic network that simultaneously embeds these three trees has at least (3/2 - ε)n reticulations. Perhaps unsurprising, caterpillar trees were also used by Baroni et al. [1] to establish that their upper bound on h(T,T') is sharp. Structurally, these trees have the property that each internal vertex is adjacent to a leaf. Each caterpillar tree can therefore be viewed as a sequence of characters, and it is exactly this viewpoint that is heavily used in [2]. More specifically, sequences with short common subsequences correspond to caterpillar trees that need many reticulations when embedded in a phylogenetic network. It would consequently be interesting to further investigate connections between caterpillar trees and certain types of sequences. Can they be used to shed more light on bounds for the minimum reticulation number?
References
[1] Baroni, M., Grünewald, S., Moulton, V., and Semple, C. (2005) "Bounding the number of hybridisation events for a consistent evolutionary history". J. Math. Biol. 51, 171–182. https://doi.org/10.1007/s00285-005-0315-9
[2] van Iersel, L., Jones, M., and Weller, M. (2023) “When three trees go to war”. HAL, ver. 3 peer-reviewed and recommended by Peer Community In Mathematical and Computational Biology. https://hal.science/hal-04013152/
DOI or URL of the preprint: https://hal.science/hal-04013152v2
Version of the preprint: 2
Dear Reviewers and Recommender,
thank you for the additional feedback. Concerning the only remaining issue, we added a phrase to the Case 2a (page 9) to make absolutely clear that the case in which all other caterpillars have no leaves mapped to leaves of L_Q is indeed included in the formulation of this case.
Just to reiterate, we are talking about the property
P(T) = caterpillar T has a leaf embedded in a leaf of L_Q
and the associated set of Caterpillars that satisfy the property:
A = { T | P(T) }
Now, Case 2a applies if all caterpillars in A have the same parity as Q. In particular, if A is empty, then the condition of Case 2a is satisfied, since all-quantified formulas over the empty set are always true. That's what I wanted to point out in my previous reviewer-answer. Admittedly, I could have been more verbose and I apologize for my previous brevity.
Dear Leo, Mark, and Mathias,
Thank you for revising your preprint `When three trees go to war’ and addressing all comments. There is one minor outstanding issue regarding the proof of Claim 1 (Case 2). Could you please have a look at the claim and address the referee's comment? If no change is necessary because the situation, as described, is already covered by Case 2a, please provide a detailed response in your reply to the comment.
Best wishes,
Simone
I am happy with the changes made to the manuscript regerding my comments.
Yet, I am not convinced by the reply to my comment 2 (about case 2a in the proof of Claim 1). My concern was the possibility of the other two caterpillars being embedded in (exactly one) leaf of N_r that is NOT a leaf of L_Q. Then the constraint “all caterpillars with a leaf embedded into a leaf of L_Q have the same parity as Q” does not apply to these caterpillars. They are not, under my assumption above, embedded in a leaf of L_Q. So unfortunately, I still don't see why this situation is covered in case 2a.
https://doi.org/10.24072/pci.mcb.100187.rev21DOI or URL of the preprint: https://hal.science/hal-04013152v1
Version of the preprint: 1
Dear Leo, Mark, and Mathias,
Thank you for submitting your preprint `When three trees go to war’ to PCI Math Comp Biol. I have received two expert reports that you can read below. Both reports comment positively on the relevance of your work, the clarity of the writing, and the impact it may have on future work in the theoretical as well as the more applied space of research on inferring phylogenetic networks from phylogenetic trees.
Before final recommendation, I ask you to please prepare a revised version of your work that addresses the referees' comments. Below, I also include a short list of additional things that I noticed while reading your paper.
I look forward to receiving your revised preprint.
Best wishes,
Simone
line 5: is $n-2$ --> is at most $n-2$
abstract: Please mention that the problem you are considering is on rooted trees and networks.
line 53: Mention briefly what a universal network is (before saying how it can be constructed).
line 75: What does `its' refer to?
line 93: unique minimum --> unique minimum node
line 118: Has your construction to find triples of caterpillars that are `as different as possible' any connections to work on strings and sequences? You already comment that the caterpillar construction problem is essentially a problem on finding strings that have short common subsequences. Is anything known about how short these subsequences can get for a fixed number of sequences that consist of the same (multi)set of letters?
page 11: Add a full stop after the first and third (centred) inequalities.
line 364: Emphasise that such a network with 2(n-2) reticulations is not necessarily most parsimonious.
Review of the manuscript: "When three trees go to war", by Leo van Iersel, Mark Jones and Mathias Weller.
In this manuscript, the authors investigate the question of the number of reticulation vertices in a network displaying a given set of $k$ phylogentic trees on $n$ leaves. For the case $k=2$, which was solved in 2005, the answer is $n-2$, meaning that for two phylogenetic trees, there always exists a network with at most $n-2$ reticulation vertices displaying both trees. In this contribution, the authors show that for $k \geq 3$, the answer is at least $(3/2-\epsilon)n$. More specifically, they show that for all $\epsilon>0$, there exists $n>0$ and three trees on $n$ leaves such that any network displaying these three trees has at least $(3/2-\epsilon)n$ reticulation vertices (Theorem 1). To the best of my knowledge, this is the first identified lower bound for this problem. Along the way, they also provide a bound to the number of leaves required in a multi-labelled tree (a kind of leaf-labelled tree in which two or more leaves may get the same label) displaying a given set of three phylogenetic trees (Corollary 1).
The paper is technical in essence, very well written, and I found no major flaw in the reasoning. There are however a few minor technicalities, which the authors may wish to address:
1 - Looking at the induction in the proofs of Lemma 1 and Lemma 2, I think you should state in the Preliminaries that a single isolated network is considered a MUL-network (or a single isolated edge, whichever you prefer). Your current definition implies that a MUL-tree necessarily has two or more leaves, which conflicts with the base case of your induction in both lemmas.
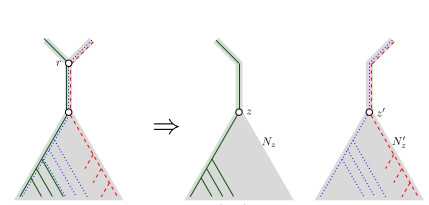
2 - In the proof of Claim 1, case 2: I am wondering about the leaves of $N_r$ that are not in $L_Q$. Should we not have a case 2a', where none of the other two caterpillars are embedded in a leaf of $L_Q$, but are both embedded in a leaf of $N_r - L_Q$? If this situation cannot happen, then the reason is not obvious to me.
3 - At the end of Rule 1, you claim that you never need to remove more token than the quantity present in the token reservoir. However, Claim 2 "only" states that the number of tokens at the end of the process is non negative. Since the proof of Claim 2 actually shows the former (stronger) statement, maybe you could rephrase Claim 2.
4 - At the very end of the proof of Claim 2. You showed that the number of bad leaves is at most $6n^{log_3 2}$, and the token reservoir contains at least $n$ tokens by the time the first withdrawal is made. But each withdrawal removes $2q$ tokens, not only $q$. So unless I am missing something, $n>6qn^{log_3 2}$ is not enough to ensure that we never remove "too many" tokens. Because with $6n^{log_3 2}$ bad leaves, we will remove $12qn^{log_3 2}$ tokens in total, not $6qn^{log_3 2}$.
5 - Some typos:
-First paragraph of section 2: "occurance" -> "occurence" (4 times).
-In Construction 1: "recusively" -> "recursively".
-In the proof of Lemma 1: "let let" -> "let".
Van Iersel, Jones, and Weller construct a family of triples of permutations that give rise to triples of rooted phylogenetic trees which are difficult to embed into a single phylogenetic network. While the problem of embedding two trees has been studied extensively, considering more trees has been regarded much harder. This manuscript yield the first lower bound on the maximum number of reticulations that can be required to embed exactly 3 binary trees.
The construction of the sequences is not very surprising, but it is hard to estimate the number of reticulations. The authors use multi-labeled trees as an intermediate structure. They first show that many leaves are needed to embed the input trees into a multi-labeled tree, and then they establish that a network with fewer reticulations than claimed would allow a multi-labeled tree with fewer leaves than possible.
The result is relevant for applications, because there is no biological reason why the embedding of k trees into a phylogenetic network should be restricted to k=2. The proofs are sophisticated and require complicated notation as well as distinguishing many cases. Some time is needed to understand the details of the proofs, but the paper is well written and organised, making it as easy as possible for the reader to follow. In the last section various questions for further research are asked which are all interesting. A natural conjecture would be that the maximum number of reticulations needed for k trees with n taxa would asymptotically be c(k)n where c(k) is the sum of 1/i for i ranging from 1 to k-1.
In summary, this is an interesting paper that is enjoyable to read and likely to have some impact on future work on the hybridization number problem.
I found four typos/minor errors that the authors might want to correct, but another round of review is not necessary.
Figure 2: Y_3 should end with 312645, not 645312.
l. 146 "let let"
ll. 236 and 243: Use 'monotonically' instead of 'monotonously'.
l.278: Add 'and' between 'that' and 'leaves'.
https://doi.org/10.24072/pci.mcb.100187.rev12