

 based on reviews by Ivan Junier and 1 anonymous reviewer
based on reviews by Ivan Junier and 1 anonymous reviewer
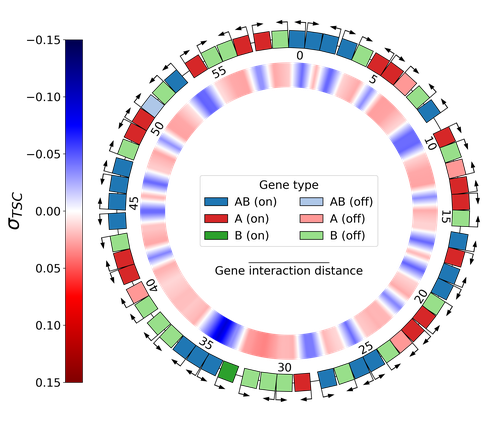
DNA supercoiling, the under or overwinding of DNA, is known to strongly impact gene expression, as changes in levels of supercoiling directly influence transcription rates. In turn, gene transcription generates DNA supercoiling on each side of an advancing RNA polymerase. This coupling between DNA supercoiling and transcription may result in different outcomes, depending on neighboring gene orientations: divergent genes tend to increase transcription levels, convergent genes tend to inhibit each other, while tandem genes may exhibit more intricate relationships.
While several works have investigated the relationship between transcription and supercoiling, Grohens et al [1] address a different question: how does transcription-supercoiling coupling drive genome evolution? To this end, they consider a simple model of gene expression regulation where transcription level only depends on the local DNA supercoiling and where the transcription of one gene generates a linear profile of positive and negative DNA supercoiling on each side of it. They then make genomes evolve through genomic inversions only considering a fitness that reflects the ability of a genome to cope with two distinct environments for which different genes have to be activated or repressed.
Using this simple model, the authors illustrate how evolutionary adaptation via genomic inversions can adjust expression levels for enhanced fitness within specific environments, particularly with the emergence of relaxation-activated genes. Investigating the genomic organization of individual genomes revealed that genes are locally organized to leverage the transcription-supercoiling coupling for activation or inhibition, but larger-scale networks of genes are required to strongly inhibit genes (sometimes up to networks of 20 genes). Thus, supercoiling-mediated interactions between genes can implicate more than just local genes. Finally, they construct an "effective interaction graph" between genes by successively simulating gene knock-outs for all of the genes of an individual and observing the effect on the expression level of other genes. They observe a densely connected interaction network, implying that supercoiling-based regulation could evolve concurrently with genome organization in bacterial genomes.
References
[1] Théotime Grohens, Sam Meyer, Guillaume Beslon (2024) Emergence of Supercoiling-Mediated Regulatory Networks through the Evolution of Bacterial Chromosome Organization. bioRxiv, ver. 4 peer-reviewed and recommended by Peer Community in Mathematical and Computational Biology https://doi.org/10.1101/2022.09.23.509185
DOI or URL of the preprint: https://doi.org/10.1101/2022.09.23.509185
Version of the preprint: 3
Dear Editor and Reviewers,
The main file containing our response to the reviewers is the reply round 2.pdf file found below.
As the edits to the main manuscript in response to the reviewers are minor, we have noted precisely in the reply file where they can be found in the main text for easy readability.
Best regards,
The authors
 , posted 07 May 2024, validated 07 May 2024
, posted 07 May 2024, validated 07 May 2024Dear Théotime, Sam, and Guillaume,
The article you have re-submitted have been reviewed a second time (see below for the two reviews). A reviewer suggests additional minor changes. We would be delighted if you would propose a new version of this preprint addressing the reviewer's comments.
I look forward to receiving your revised preprint, upon which I will submit my recommendation.
Best wishes,
Nelle
After carefully reading the modifications provided by the authors, the manuscript has been extended by incorporating the impact of gene triplets into the analysis. Following these revisions, the manuscript now presents a clearer communication of the main findings and the methods employed.
Therefore, I have no further requirements for the manuscript and am pleased to proceed with its publication.
Regarding the additional questions:
DOI or URL of the preprint: https://doi.org/10.1101/2022.09.23.509185
Version of the preprint: 2
Dear Editor and Reviewers,
The main file containing our response to the reviewers is the reply.pdf file found below.
We apologize for not providing a tracked changes document, as we have made changes throughout the manuscript that we feel would be impractical to follow in such a way. We have therefore uploaded the full manuscript in that place.
The main addition to the manuscript is a new figure (Figure 6) and the text that describes the figure, in Section 2.2, and the rest of the modifications consists in localized edits for precision or clarification.
Best regards,
The authors
 , posted 07 Dec 2023, validated 07 Dec 2023
, posted 07 Dec 2023, validated 07 Dec 2023Dear Théotime, Sam, and Guillaume,
Thank you for submitting your preprint "Emergence of Supercoiling-Mediated
Regulatory Networks through Bacterial Chromosome Organization" to PCI Math
Comp Bio. I apologize in the delay in giving you feedback.
Below, you'll find two expert reports I've received. Both reports comment
positively about the relevance of your work, the clarity of your writing, and
the high impact it has in understanding the coupling of transcription and
supercoiling. However, they point out a number of possible improvements and
raise various questions on your work.
We would be delighted if you would propose a new version of this preprint
addressing the referees' comments.
I look forward to receiving your revised preprint.
Best wishes,
Nelle
This is an interesting study where the authors investigate the impact of the coupling between DNA supercoiling and DNA transcription on the evolution of bacterial genome organization. This investigation is motivated by previous evidence suggesting that supercoiling can regulate gene expression, which, in turn, is directly linked to gene organization.
To explore this effect, the authors present a model of the transcription-supercoiling coupling. This model computes gene expression levels based on gene positions and directions in the genome. The model is integrated into an evolutionary framework where individuals can undergo mutations (gene inversions) to maximize a fitness function dependent on gene expression levels. Individuals must adapt their gene expression levels according to two environments with different superhelical levels, each requiring genes to be expressed, repressed, or both (resulting in three gene types).
The results of this study highlights the importance of gene organization and its direct impact on gene expression levels, driven by DNA supercoiling. These interactions extend beyond pairwise interactions between neighboring genes, revealing a complex web of connections.
Overall, the article is well-written and effectively communicates the main findings and methods.
Here are some minor suggestions to enhance the quality and clarity of the paper:
1. In the abstract, "two different environments" are mentioned without immediate context. After reading the paper, now I know what it means, but it would be beneficial to clarify that these two environments refer to settings with different global superhelical levels. This clarification will help readers better understand the specific conditions being discussed.
2. While the introduction effectively presents the research question, which investigates the impact of transcription-supercoiling coupling on genome evolution in bacteria, consider adding a more explicit discussion of the potential broader implications and significance of answering this research question. Mentioning how this research could advance the field, benefit experimentalists, contribute to synthetic biology, inform future models, etc., would provide a clearer picture of the research's potential impact.
3. Explicitly mention the mathematical objective of the model in the methods section (I didn't see it there). Specifically, state that the model aims to maximize the fitness function 'f' based on gene type assignments (A, B, AB). Including this clarification will provide a clearer understanding of the primary mathematical goal and purpose of the model within the methods section.
4. I found it somewhat unclear from the text whether, for each generation, there were N=100 copies of individuals with the same mutation or if random mutations were introduced for each of the 100 individuals. While the latter interpretation seems more reasonable to me, explicitly mentioning this in the text would greatly enhance clarity and help readers better understand the experimental setup.
5. For Figures 5, 7, 9 & 10, explicitly indicate the significance of the circular markers 'o' in the captions. ' It might not be immediately clear to readers what these markers represent, and including this information in the captions would facilitate a better understanding of the data and visuals.
6. Mention the threshold used to define the minimal subnetwork size (around line 325). Additionally, it would be helpful to have figures depicting the expression levels of both the minimal subnetworks and the complete genome. Perhaps these figures could be included as supplementary material to provide a more comprehensive view of the data.
7. Revise the captions of Figure 10 to explicitly explain and mention the box plots, ensuring that each figure is understandable on its own.
8. In line 603, where it states "n= 1 if the genes are on the same strand, and n=-1 otherwise" shouldn't it be the other strand?
9. In Section 5.3 (Experimental Setup), include a reminder that each individual is tested in the two environments.
10. In the same section (5.3 Experimental Setup), if I understood correctly, the mathematical objective is to maximize the fitness function 'f' based on the assigned gene types. It could be beneficial to explicitly state this objective in the methods section for clarity.
11. Overall, the methods are well-presented, but the addition of a diagram illustrating the simulation process might enhance the comprehensibility of the procedure, making it easier for readers to visualize the steps involved.