Latest recommendations
| Id | Title * | Authors * | Abstract * | Picture * | Thematic fields * | Recommender | Reviewers | Submission date | |
|---|---|---|---|---|---|---|---|---|---|
22 Apr 2025
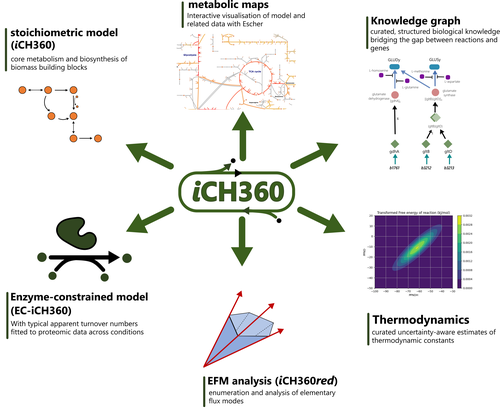
A compact model of Escherichia coli core and biosynthetic metabolismMarco Corrao, Hai He, Wolfram Liebermeister, Elad Noor, Arren Bar-Even https://doi.org/10.48550/arXiv.2406.16596‘Goldilocks’-size extensively annotated model for Escherichia coli metabolismRecommended by Meike Wortel based on reviews by Daan de Groot, Benjamin Luke Coltman and 1 anonymous reviewer based on reviews by Daan de Groot, Benjamin Luke Coltman and 1 anonymous reviewer
Metabolism is the driving force of life and thereby plays a key role in understanding microbial functioning in monoculture and in ecosystems, from natural habitats to biotechnological applications, from microbiomes related to human health to food production. However, the complexity of metabolic networks poses a major challenge for understanding how they are shaped by evolution and how we can manipulate them. Therefore, many network-based methods have been developed to study metabolism. On the other end are well-curated small-scale models of metabolic pathways. For those, knowledge of the enzymes of a pathway, their kinetic properties and (optionally) regulation by metabolites is incorporated in usually a differential equation model. Standard methods for systems of differential equations can be used to study steady-states and the dynamics of these models, which can lead to accurate predictions (Flamholz et al., 2013; van Heerden et al., 2014). However, the downside is that the methods are difficult to scale up and, for many enzymes, the detailed information necessary for these models is not available. Combined with computational challenges, these models are limited to specific pathways and cannot be used for whole cells, nor even communities. Therefore, there is still a need for both methods and models to make accurate predictions on a scale beyond single pathways. Corrao et al. (2025) aim for an intermediate size model that is both accurate and predictive, does not need an extensive set of enzyme parameters, but also encompasses most of the cell’s metabolic pathways. As they phrase it: a model in the ‘Goldilocks’ zone. Curation can improve genome-scale models substantially but requires additional experimental data. However, as the authors show, even the well-curated model of Escherichia coli can sometimes show unrealistic metabolic flux patterns. A smaller model can be better curated and therefore more predictive, and more methods can be applied, as for example EFM based approaches. The authors show an extensive set of methodologies that can be applied to this model and yield interpretable results. Additionally, the model contains a wealth of standardized annotation that could set a standard for the field. This is a first model of its kind, and it is not surprising that E. coli is used as its metabolism is very well-studied. However, this could set the basis for similar models for other well-studied organisms. Because the model is well-annotated and characterized, it is very suitable for testing new methods that make predictions with such an intermediate-sized model and that can later be extended for larger models. In the future, such models for different species could aid the creation of methods for studying and predicting metabolism in communities, for which there is a large need for applications (e.g. bioremediation and human health). The different layers of annotation and the available code with clear documentation make this model an ideal resource as teaching material as well. Methods can be explained on this model, which can still be visualized and interpreted because of its reduced size, while it is large enough to show the differences between methods. Although it might be too much to expect models of this type for all species, the different layers of annotation can be used to inspire better annotation of genome-scale models and enhance their accuracy and predictability. Thus, this paper sets a standard that could benefit research on metabolic pathways from individual strains to natural communities to communities for biotechnology, bioremediation and human health. References Bauer, E., Zimmermann, J., Baldini, F., Thiele, I., Kaleta, C., 2017. BacArena: Individual-based metabolic modeling of heterogeneous microbes in complex communities. PLOS Comput. Biol. 13, e1005544. https://doi.org/10.1371/journal.pcbi.1005544 Corrao, M., He, H., Liebermeister, W., Noor, E., Bar-Even, A., 2025. A compact model of Escherichia coli core and biosynthetic metabolism. https://doi.org/10.48550/arXiv.2406.16596 Dukovski, I., Bajić, D., Chacón, J.M., Quintin, M., Vila, J.C.C., Sulheim, S., Pacheco, A.R., Bernstein, D.B., Riehl, W.J., Korolev, K.S., Sanchez, A., Harcombe, W.R., Segrè, D., 2021. A metabolic modeling platform for the computation of microbial ecosystems in time and space (COMETS). Nat. Protoc. 16, 5030–5082. https://doi.org/10.1038/s41596-021-00593-3 Flamholz, A., Noor, E., Bar-Even, A., Liebermeister, W., Milo, R., 2013. Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc. Natl. Acad. Sci. 110, 10039–10044. https://doi.org/10.1073/pnas.1215283110 Gralka, M., Pollak, S., Cordero, O.X., 2023. Genome content predicts the carbon catabolic preferences of heterotrophic bacteria. Nat. Microbiol. 8, 1799–1808. https://doi.org/10.1038/s41564-023-01458-z Henry, C.S., DeJongh, M., Best, A.A., Frybarger, P.M., Linsay, B., Stevens, R.L., 2010. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28, 977–982. https://doi.org/10.1038/nbt.1672 Li, Z., Selim, A., Kuehn, S., 2023. Statistical prediction of microbial metabolic traits from genomes. PLOS Comput. Biol. 19, e1011705. https://doi.org/10.1371/journal.pcbi.1011705 Machado, D., Andrejev, S., Tramontano, M., Patil, K.R., 2018. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 46, 7542–7553. https://doi.org/10.1093/nar/gky537 Mendoza, S.N., Olivier, B.G., Molenaar, D., Teusink, B., 2019. A systematic assessment of current genome-scale metabolic reconstruction tools. Genome Biol. 20, 158. https://doi.org/10.1186/s13059-019-1769-1 Orth, J.D., Thiele, I., Palsson, B.Ø., 2010. What is flux balance analysis? Nat. Biotechnol. 28, 245–248. https://doi.org/10.1038/nbt.1614 Scott Jr, W.T., Benito-Vaquerizo, S., Zimmermann, J., Bajić, D., Heinken, A., Suarez-Diez, M., Schaap, P.J., 2023. A structured evaluation of genome-scale constraint-based modeling tools for microbial consortia. PLOS Comput. Biol. 19, e1011363. https://doi.org/10.1371/journal.pcbi.1011363 van Heerden, J.H., Wortel, M.T., Bruggeman, F.J., Heijnen, J.J., Bollen, Y.J.M., Planqué, R., Hulshof, J., O’Toole, T.G., Wahl, S.A., Teusink, B., 2014. Lost in Transition: Start-Up of Glycolysis Yields Subpopulations of Nongrowing Cells. Science 343, 1245114. https://doi.org/10.1126/science.1245114 | A compact model of Escherichia coli core and biosynthetic metabolism | Marco Corrao, Hai He, Wolfram Liebermeister, Elad Noor, Arren Bar-Even | <p>Metabolic models condense biochemical knowledge about organisms in a structured and standardised way. As large-scale network reconstructions are readily available for many organisms, genome-scale models are being widely used among modellers and... |  | Cell Biology, Systems biology | Meike Wortel | 2024-10-22 10:26:48 | View | |
30 Mar 2025
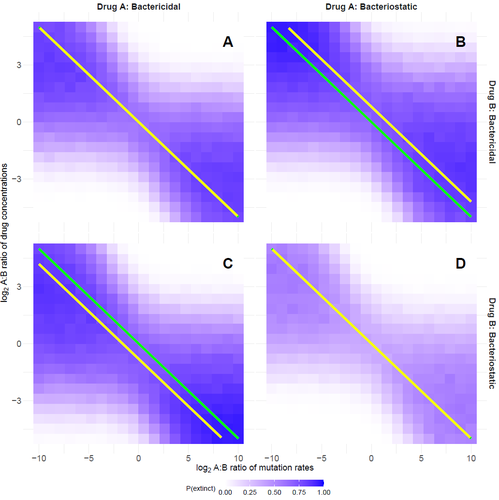
Optimal antimicrobial dosing combinations when drug-resistance mutation rates differOscar Delaney, Andrew D. Letten, Jan Engelstaedter https://doi.org/10.1101/2024.05.04.592498Optimizing antibiotic use: How much should we favor drugs with lower resistance mutation rates?Recommended by Amaury Lambert based on reviews by 2 anonymous reviewersIn the hereby recommended paper [1], Delaney, Letten and Engelstädter study the appearance of antibiotic resistance(s) in a bacterial population subject to a combination of two antibiotics \( A \) and \( B \), in concentrations \( C_A \) and \( C_B \) respectively. Their goal was to find optimal values of \( C_A \) and \( C_B \) that minimize the risk of evolutionary rescue, under the unusual assumption that resistance mutations to either antibiotic are not equally likely. The authors introduce a stochastic model assuming that the susceptible population grows like a supercritical birth-death process which becomes subcritical in the presence of antibiotics (and exactly critical for a certain concentration c of a single antibiotic): the effect of each antibiotic is either to reduce division rate (bacteriostatic drug) or to enhance death rate (bacteriocidal drug) by a factor which has a sigmoid functional dependence on antibiotic concentration. Now at each division, a resistance mutation can arise, with probability \( \mu_A \) for a resistance to antibiotic \( A \) and with probability \( \mu_B \) for a resistance to antibiotic \( B \). The goal of the paper is to find the optimal ratio of drug concentrations \( C_A \) and \( C_B \) when these are subject to a constrain \( C_A + C_B = c \), depending on the ratio of mutation rates. Assuming total resistance and no cross resistance, the authors show that the optimal concentrations are given by \( C_A = c/(1+\sqrt{\mu_A/\mu_B }) \) and \( C_B = c/(1+\sqrt{\mu_B/\mu_A}) \), which leads to the beautiful result that the optimal ratio \( C_A/C_B \) is equal to the square root of \( \mu_B/\mu_A \). The authors have made a great job completing their initial submission by simulations of model extensions, relaxing assumptions like single antibiotic mode, absence of competition, absence of cost of resistance, sharp cutoff in toxicity… and comparing the results obtained by simulation to their mathematical result. The paper is very clearly written and any reader interested in antibiotic resistance, stochastic modeling of bacterial populations and/or evolutionary rescue will enjoy reading it. Let me thank the authors for their patience and for their constant willingness to comply with the reviewers’ and recommender’s demands during the reviewing process.
Reference Oscar Delaney, Andrew D. Letten, Jan Engelstaedter (2025) Optimal antimicrobial dosing combinations when drug-resistance mutation rates differ. bioRxiv, ver.3 peer-reviewed and recommended by PCI Mathematical and Computational Biology https://doi.org/10.1101/2024.05.04.592498 | Optimal antimicrobial dosing combinations when drug-resistance mutation rates differ | Oscar Delaney, Andrew D. Letten, Jan Engelstaedter | <p>Given the ongoing antimicrobial resistance crisis, it is imperative to develop dosing regimens optimised to avoid the evolution of resistance. The rate at which bacteria acquire resistance-conferring mutations to different antimicrobial drugs s... |  | Evolutionary Biology | Amaury Lambert | 2024-05-07 17:17:55 | View | |
25 Feb 2025
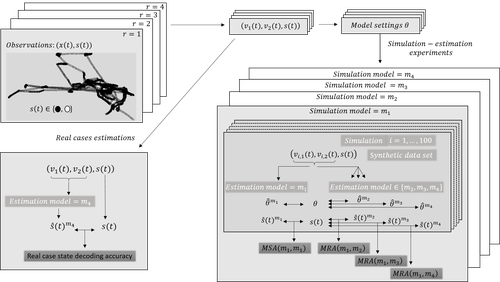
Proper account of auto-correlations improves decoding performances of state-space (semi) Markov modelsNicolas Bez, Pierre Gloaguen, Marie-Pierre Etienne, Rocio Joo, Sophie Lanco, Etienne Rivot, Emily Walker, Mathieu Woillez, Stéphanie Mahévas https://hal.science/hal-04547315An empirical study on the impact of neglecting dependencies in the observed or the hidden layer of a H(S)MM model on decoding performancesRecommended by Nathalie Peyrard based on reviews by Sandra Plancade and 1 anonymous reviewer based on reviews by Sandra Plancade and 1 anonymous reviewer
The article by Bez et al [1] addresses an important issue for statisticians and ecological modellers: the impact of modelling choices when considering state-space models to represent time series with hidden regimes. The authors present an empirical study of the impact of model misspecification for models in the HMM and HSMM family. The misspecification can be at the level of the hidden chain (Markovian or semi-Markovian assumption) or at the level of the observed chain (AR0 or AR1 assumption). The study uses data on the movements of fishing vessels. Vessels can exert pressure on fish stocks when they are fishing, and the aim is to identify the periods during which fishing vessels are fishing or not fishing, based on GPS tracking data. Two sets of data are available, from two vessels with contrasting fishing behaviour. The empirical study combines experiments on the two real datasets and on data simulated from models whose parameters are estimated on the real datasets. In both cases, the actual sequence of activities is available. The impact of a model misspecification is mainly evaluated on the restored hidden chain (decoding task), which is very relevant since in many applications we are more interested in the quality of decoding than in the accuracy of parameters estimation. Results on parameter estimation are also presented and metrics are developed to help interpret the results. The study is conducted in a rigorous manner and extensive experiments are carried out, making the results robust. The main conclusion of the study is that choosing the wrong AR model at the observed sequence level has more impact than choosing the wrong model at the hidden chain level. The article ends with an interesting discussion of this finding, in particular the impact of resolution on the quality of the decoding results. As the authors point out in this discussion, the results of this study are not limited to the application of GPS data to the activities of fishing vessels Beyond ecology, H(S)MMs are also widely used epidemiology, seismology, speech recognition, human activity recognition ... The conclusion of this study will therefore be useful in a wide range of applications. It is a warning that should encourage modellers to design their hidden Markov models carefully or to interpret their results cautiously. References [1] Nicolas Bez, Pierre Gloaguen, Marie-Pierre Etienne, Rocio Joo, Sophie Lanco, Etienne Rivot, Emily Walker, Mathieu Woillez, Stéphanie Mahévas (2024) Proper account of auto-correlations improves decoding performances of state-space (semi) Markov models. HAL, ver.3 peer-reviewed and recommended by PCI Math Comp Biol https://hal.science/hal-04547315v3 | Proper account of auto-correlations improves decoding performances of state-space (semi) Markov models | Nicolas Bez, Pierre Gloaguen, Marie-Pierre Etienne, Rocio Joo, Sophie Lanco, Etienne Rivot, Emily Walker, Mathieu Woillez, Stéphanie Mahévas | <p>State-space models are widely used in ecology to infer hidden behaviors. This study develops an extensive numerical simulation-estimation experiment to evaluate the state decoding accuracy of four simple state-space models. These models are obt... |  | Dynamical systems, Ecology, Probability and statistics | Nathalie Peyrard | 2024-05-29 16:29:25 | View | |
27 Jan 2025

Biology-Informed inverse problems for insect pests detection using pheromone sensorsThibault Malou, Nicolas Parisey, Katarzyna Adamczyk-Chauvat, Elisabeta Vergu, Béatrice Laroche, Paul-Andre Calatayud, Philippe Lucas, Simon Labarthe https://hal.inrae.fr/hal-04572831v2Towards accurate inference of insect presence landscapes from pheromone sensor networksRecommended by Eric Tannier based on reviews by Angelo Iollo and 1 anonymous reviewer based on reviews by Angelo Iollo and 1 anonymous reviewer
Insecticides are used to control crop pests and prevent severe crop losses. They are also a major cause of the current decline in biodiversity, contribute to climate change, and pollute soil and water, with consequences for human and environmental health [1]. The rationale behind the work of Malou et al [2] is that some pesticide application protocols can be improved by a better knowledge of the insects, their biology, their ecology and their real-time infestation dynamics in the fields. Thanks to a network of pheromone sensors and a mathematical method to derive the spatio-temporal distribution of pest populations from the signals, it is theoretically possible to adjust the time, dose and area of treatment and to use less pesticide with greater efficiency than an uninformed protocol. Malou et al [2] focus on the mathematical problem, recognising that its real role in pest control would require work on its implementation and on a benefit-harm analysis. The problem is an "inverse problem" [3] in that it consists of inferring the presence of insects from the trail left by the pheromones, given a model of pheromone diffusion by insects. The main contribution of this work is the formulation and comparison of different regularisation terms in the optimisation inference scheme, in order to guide the optimisation by biological knowledge of specific pests, such as some parameters of population dynamics. The accuracy and precision of the results are tested and compared on a simple toy example to test the ability of the model and algorithm to detect the source of the pheromones and the efficiency of the data assimilation principle. A further simulation is then carried out on a real plot with realistic parameters and rules based on knowledge of a maize pest. A repositioning of the sensors (informed by the results from the initial positions) is carried out during the test phase to allow better detection. The work of Malou et al [2] is large, deep and complete. Its includes a detailed study of the numerical solutions of different data assimilation methods, as well as a theoretical reflection on how this work could contribute to agricultural and environmental issues. References [1] IPBES (2024). Thematic Assessment Report on the Underlying Causes of Biodiversity Loss and the Determinants of Transformative Change and Options for Achieving the 2050 Vision for Biodiversity of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services. O’Brien, K., Garibaldi, L., and Agrawal, A. (eds.). IPBES secretariat, Bonn, Germany. https://doi.org/10.5281/zenodo.11382215 [2] Thibault Malou, Nicolas Parisey, Katarzyna Adamczyk-Chauvat, Elisabeta Vergu, Béatrice Laroche, Paul-Andre Calatayud, Philippe Lucas, Simon Labarthe (2025) Biology-Informed inverse problems for insect pests detection using pheromone sensors. HAL, ver.2 peer-reviewed and recommended by PCI Math Comp Biol https://hal.inrae.fr/hal-04572831v2 [3] Isakov V (2017). Inverse Problems for Partial Differential Equations. Vol. 127. Applied Mathematical Sciences. Cham: Springer International Publishing. https://doi.org/10.1007/978-3-319-51658-5. | Biology-Informed inverse problems for insect pests detection using pheromone sensors | Thibault Malou, Nicolas Parisey, Katarzyna Adamczyk-Chauvat, Elisabeta Vergu, Béatrice Laroche, Paul-Andre Calatayud, Philippe Lucas, Simon Labarthe | <p>Most insects have the ability to modify the odor landscape in order to communicate with their conspecies during key phases of their life cycle such as reproduction. They release pheromones in their nearby environment, volatile compounds that ar... |  | Agricultural Science, Dynamical systems, Epidemiology, Systems biology | Eric Tannier | 2024-05-12 19:14:34 | View | |
08 Nov 2024

Bayesian joint-regression analysis of unbalanced series of on-farm trialsMichel Turbet Delof , Pierre Rivière , Julie C Dawson, Arnaud Gauffreteau , Isabelle Goldringer , Gaëlle van Frank , Olivier David https://hal.science/hal-04380787Handling Data Imbalance and G×E Interactions in On-Farm Trials Using Bayesian Hierarchical ModelsRecommended by Sophie Donnet based on reviews by Pierre Druilhet and David Makowski based on reviews by Pierre Druilhet and David Makowski
The article, "Bayesian Joint-Regression Analysis of Unbalanced Series of On-Farm Trials," presents a Bayesian statistical framework tailored for analyzing highly unbalanced datasets from participatory plant breeding (PPB) trials, specifically wheat trials. The key goal of this research is to address the challenges of genotype-environment (G×E) interactions in on-farm trials, which often have limited replication and varied testing conditions across farms. The study applies a hierarchical Bayesian model with Finlay-Wilkinson regression, which improves the estimation of G×E effects despite substantial data imbalance. By incorporating a Student’s t-distribution for residuals, the model is more robust to extreme values, which are common in on-farm trials due to variable environments. Note that the model allows a detailed breakdown of variance, identifying environment effects as the most significant contributors, thus highlighting areas for future breeding focus. Using Hamiltonian Monte Carlo methods, the study achieves reasonable computation times, even for large datasets. Obviously, the limitation of the methods comes from the level of data balance and replication. The method requires a minimum level of data balance and replication, which can be a challenge in very decentralized breeding networks Moreover, the Bayesian framework, though computationally feasible, may still be complex for widespread adoption without computational resources or statistical expertise. The paper presents a sophisticated Bayesian framework specifically designed to tackle the challenges of unbalanced data in participatory plant breeding (PPB). It showcases a novel way to manage the variability in on-farm trials, a common issue in decentralized breeding programs. This study's methods accommodate the inconsistencies inherent in on-farm trials, such as extreme values and minimal replication. By using a hierarchical Bayesian approach with a Student’s t-distribution for robustness, it provides a model that maintains precision despite these real-world challenges. This makes it especially relevant for those working in unpredictable agricultural settings or decentralized trials. From a more general perspective, this paper’s findings support breeding methods that prioritize specific adaptation to local conditions. It is particularly useful for researchers and practitioners interested in breeding for agroecological or organic farming systems, where G×E interactions are critical but hard to capture in traditional trial setups. Beyond agriculture, the paper serves as an excellent example of advanced statistical modeling in highly variable datasets. Its applications extend to any field where data is incomplete or irregular, offering a clear case for hierarchical Bayesian methods to achieve reliable results. Finally, although begin quite methodological, the paper provides practical insights into how breeders and researchers can work with farmers to achieve meaningful varietal evaluations. References Michel Turbet Delof , Pierre Rivière , Julie C Dawson, Arnaud Gauffreteau , Isabelle Goldringer , Gaëlle van Frank , Olivier David (2024) Bayesian joint-regression analysis of unbalanced series of on-farm trials. HAL, ver.2 peer-reviewed and recommended by PCI Math Comp Biol https://hal.science/hal-04380787 | Bayesian joint-regression analysis of unbalanced series of on-farm trials | Michel Turbet Delof , Pierre Rivière , Julie C Dawson, Arnaud Gauffreteau , Isabelle Goldringer , Gaëlle van Frank , Olivier David | <p>Participatory plant breeding (PPB) is aimed at developing varieties adapted to agroecologically-based systems. In PPB, selection is decentralized in the target environments, and relies on collaboration between farmers, farmers' organisations an... |  | Agricultural Science, Genetics and population Genetics, Probability and statistics | Sophie Donnet | Pierre Druilhet, David Makowski | 2024-01-11 14:17:41 | View |
21 Oct 2024
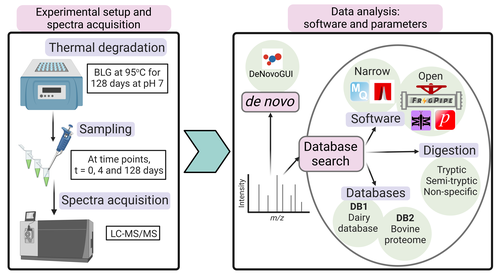
Benchmarking the identification of a single degraded protein to explore optimal search strategies for ancient proteinsIsmael Rodriguez-Palomo, Bharath Nair, Yun Chiang, Joannes Dekker, Benjamin Dartigues, Meaghan Mackie, Miranda Evans, Ruairidh Macleod, Jesper V. Olsen, Matthew J. Collins https://doi.org/10.1101/2023.12.15.571577Systematic investigation of software tools and design of a tailored pipeline for paleoproteomics researchRecommended by Raquel Assis based on reviews by Shevan Wilkin and 1 anonymous reviewerPaleoproteomics is a rapidly growing field with numerous challenges, many of which are due to the highly fragmented, modified, and degraded nature of ancient proteins. Though there are established standards for analysis, it is unclear how different software tools affect the identification and quantification of peptides, proteins, and post-translational modifications. To address this knowledge gap, Rodriguez Palomo et al. design a controlled system by experimentally degrading and purifying bovine beta-lactoglobulin, and then systematically compare the performance of many commonly used tools in its analysis. They present comprehensive investigations of false discovery rates, open and narrow searches, de novo sequencing coverage bias and accuracy, and peptide chemical properties and bias. In each investigation, they explore wide ranges of appropriate tools and parameters, providing guidelines and recommendations for best practices. Based on their findings, Rodriguez Palomo et al. develop a proposed pipeline that is tailored for the analysis of ancient proteins. This pipeline is an important contribution to paleoproteomics and is likely to be of great value to the research community, as it is designed to enhance power, accuracy, and consistency in studies of ancient proteins. References Ismael Rodriguez-Palomo, Bharath Nair, Yun Chiang, Joannes Dekker, Benjamin Dartigues, Meaghan Mackie, Miranda Evans, Ruairidh Macleod, Jesper V. Olsen, Matthew J. Collins (2023) Benchmarking the identification of a single degraded protein to explore optimal search strategies for ancient proteins. bioRxiv, ver.3 peer-reviewed and recommended by PCI Math Comp Biol https://doi.org/10.1101/2023.12.15.571577 | Benchmarking the identification of a single degraded protein to explore optimal search strategies for ancient proteins | Ismael Rodriguez-Palomo, Bharath Nair, Yun Chiang, Joannes Dekker, Benjamin Dartigues, Meaghan Mackie, Miranda Evans, Ruairidh Macleod, Jesper V. Olsen, Matthew J. Collins | <p style="text-align: justify;">Palaeoproteomics is a rapidly evolving discipline, and practitioners are constantly developing novel strategies for the analyses and interpretations of complex, degraded protein mixtures. The community has also esta... |  | Genomics and Transcriptomics, Probability and statistics | Raquel Assis | Anonymous, Shevan Wilkin | 2024-03-12 15:17:08 | View |
02 Oct 2024
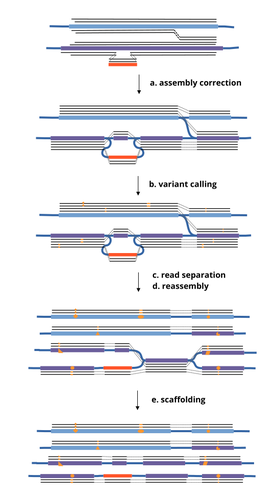
HairSplitter: haplotype assembly from long, noisy readsRoland Faure, Dominique Lavenier, Jean-François Flot https://doi.org/10.1101/2024.02.13.580067Accurate Haplotype Reconstruction from Long, Error-Prone, Reads with HairSplitterRecommended by Giulio Ermanno Pibiri based on reviews by Dmitry Antipov and 1 anonymous reviewer based on reviews by Dmitry Antipov and 1 anonymous reviewer
A prominent challenge in computational biology is to distinguish microbial haplotypes -- closely related organisms with highly similar genomes -- due to small genomic differences that can cause significant phenotypic variations. Current genome assembly tools struggle with distinguishing these haplotypes, especially for long-read sequencing data with high error rates, such as PacBio or Oxford Nanopore Technology (ONT) reads. While existing methods work well for either viral or bacterial haplotypes, they often fail with low-abundance haplotypes and are computationally intensive. This work by Faure, Lavenier, and Flot [1] introduces a new tool -- HairSplitter -- that offers a solution for both viral and bacterial haplotype separation, even with error-prone long reads. It does this by efficiently calling variants, clustering reads into haplotypes, creating new separated contigs, and resolving the assembly graph. A key advantage of HairSplitter is that it is entirely parameter-free and does not require prior knowledge of the organism's ploidy. HairSplitter is designed to handle both metaviromes and bacterial metagenomes, offering a more versatile and efficient solution than existing tools, like stRainy [2], Strainberry [3], and hifiasm-meta [4]. References [1] Roland Faure, Dominique Lavenier, Jean-François Flot (2024) HairSplitter: haplotype assembly from long, noisy reads. bioRxiv, ver.3 peer-reviewed and recommended by PCI Math Comp Biol https://doi.org/10.1101/2024.02.13.580067 [2] Kazantseva E, A Donmez, M Pop, and M Kolmogorov (2023). stRainy: assembly-based metagenomic strain phasing using long reads. Bioinformatics. https://doi.org/10.1101/2023.01.31.526521 [3] Vicedomini R, C Quince, AE Darling, and R Chikhi (2021). Strainberry: automated strain separation in low complexity metagenomes using long reads. Nature Communications, 12, 4485. ISSN: 2041-1723. https://doi.org/10.1038/s41467-021-24515-9 [4] Feng X, H Cheng, D Portik, and H Li (2022). Metagenome assembly of high-fidelity long reads with hifiasm-meta. Nature Methods, 19, 1–4. https://doi.org/10.1038/s41592-022-01478-3 | HairSplitter: haplotype assembly from long, noisy reads | Roland Faure, Dominique Lavenier, Jean-François Flot | <p>Long-read assemblers face challenges in discerning closely related viral or<br>bacterial strains, often collapsing similar strains in a single sequence. This limitation has<br>been hampering metagenome analysis, where diverse strains may harbor... |  | Design and analysis of algorithms, Development, Genomics and Transcriptomics, Probability and statistics | Giulio Ermanno Pibiri | 2024-02-15 10:17:04 | View | |
27 Sep 2024
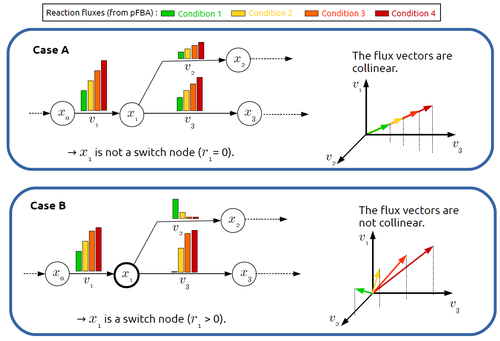
In silico identification of switching nodes in metabolic networksFrancis Mairet https://doi.org/10.1101/2023.05.17.541195A computational method to identify key players in metabolic rewiringRecommended by Claudine Chaouiya based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
Significant progress has been made in developing computational methods to tackle the analysis of the numerous (genome-wide scale) metabolic networks that have been documented for a wide range of species. Understanding the behaviours of these complex reaction networks is crucial in various domains such as biotechnology and medicine. Metabolic rewiring is essential as it enables cells to adapt their metabolism to changing environmental conditions. Identifying the metabolites around which metabolic rewiring occurs is certainly useful in the case of metabolic engineering, which relies on metabolic rewiring to transform micro-organisms into cellular factories [1], as well as in other contexts. This paper by F. Mairet [2] introduces a method to disclose these metabolites, named switch nodes, relying on the analysis of the flux distributions for different input conditions. Basically, considering fluxes for different inputs, which can be computed using e.g. Parsimonious Flux Balance Analysis (pFBA), the proposed method consists in identifying metabolites involved in reactions whose different flux vectors are not collinear. The approach is supported by four case studies, considering core and genome-scale metabolic networks of Escherichia coli, Saccharomyces cerevisiae and the diatom Phaeodactylum tricornutum. Whilst identified switch nodes may be biased because computed flux vectors satisfying given objectives are not necessarily unique, the proposed method has still a relevant predictive potential, complementing the current array of computational methods to study metabolism. References [1] Tao Yu, Yasaman Dabirian, Quanli Liu, Verena Siewers, Jens Nielsen (2019) Strategies and challenges for metabolic rewiring. Current Opinion in Systems Biology, Vol 15, pp 30-38. https://doi.org/10.1016/j.coisb.2019.03.004. [2] Francis Mairet (2024) In silico identification of switching nodes in metabolic networks. bioRxiv, ver.3 peer-reviewed and recommended by PCI Math Comp Biol https://doi.org/10.1101/2023.05.17.541195 | In silico identification of switching nodes in metabolic networks | Francis Mairet | <p>Cells modulate their metabolism according to environmental conditions. A major challenge to better understand metabolic regulation is to identify, from the hundreds or thousands of molecules, the key metabolites where the re-orientation of flux... |  | Graph theory, Physiology, Systems biology | Claudine Chaouiya | Anonymous | 2023-05-26 17:24:26 | View |
27 Aug 2024
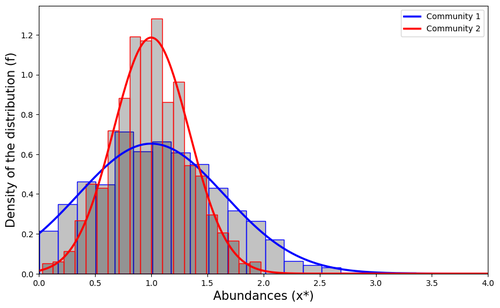
Impact of a block structure on the Lotka-Volterra modelMaxime Clenet, François Massol, Jamal Najim https://doi.org/10.48550/arXiv.2311.09470Equlibrium of communities in the Lotka-Volterra modelRecommended by Loïc Paulevé based on reviews by 3 anonymous reviewers based on reviews by 3 anonymous reviewers
This article by Clenet et al. [1] tackles a fundamental mathematical model in ecology to understand the impact of the architecture of interactions on the equilibrium of the system. The authors consider the classical Lotka-Volterra model, depicting the effect of interactions between species on their abundances. They focus on the case whenever there are numerous species, and where their interactions are compartmentalized in a block structure. Each block has a strength coefficient, applied to a random Gaussian matrix. This model aims at capturing the structure of interacting communities, with blocks describing the interactions within a community, and other blocks the interactions between communities. In this general mathematical framework, the authors demonstrate sufficient conditions for the existence and uniqueness of a stable equilibrium, and conditions for which the equilibrium is feasible. Moreover, they derive statistical heuristics for the proportion, mean, and distribution of abundance of surviving species. Overall, the article constitutes an original and solid contribution to the study of mathematical models in ecology. It combines mathematical analysis, dynamical system theory, numerical simulations, grounded with relevant hypothesis for the modeling of ecological systems. References [1] Maxime Clenet, François Massol, Jamal Najim (2023) Impact of a block structure on the Lotka-Volterra model. arXiv, ver.3 peer-reviewed and recommended by Peer Community in Mathematical and Computational Biology. https://doi.org/10.48550/arXiv.2311.09470 | Impact of a block structure on the Lotka-Volterra model | Maxime Clenet, François Massol, Jamal Najim | <p>The Lotka-Volterra (LV) model is a simple, robust, and versatile model used to describe large interacting systems such as food webs or microbiomes. The model consists of $n$ coupled differential equations linking the abundances of $n$ differen... |  | Dynamical systems, Ecology, Probability and statistics | Loïc Paulevé | 2023-11-17 21:44:38 | View | |
13 Aug 2024
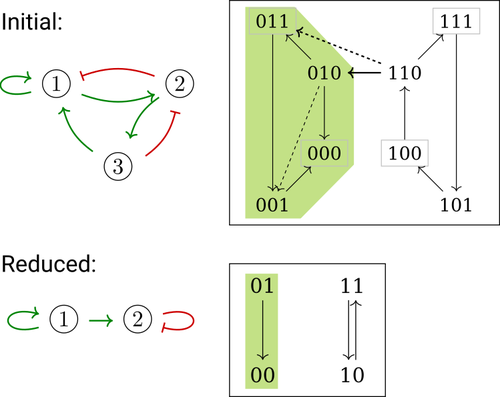
Phenotype control and elimination of variables in Boolean networksElisa Tonello, Loïc Paulevé https://doi.org/10.48550/arXiv.2406.02304Disclosing effects of Boolean network reduction on dynamical properties and control strategiesRecommended by Claudine Chaouiya based on reviews by Tomas Gedeon and David Safranek based on reviews by Tomas Gedeon and David Safranek
Boolean networks stem from seminal work by M. Sugita [1], S. Kauffman [2] and R. Thomas [3] over half a century ago. Since then, a very active field of research has been developed, leading to theoretical advances accompanied by a wealth of work on modelling genetic and signalling networks involved in a wide range of cellular processes. Boolean networks provide a successful formalism for the mathematical modelling of biological processes, with a qualitative abstraction particularly well adapted to handle the modelling of processes for which precise, quantitative data is barely available. Nevertheless, these abstract models reveal fundamental dynamical properties, such as the existence and reachability of attractors, which embody stable cellular responses (e.g. differentiated states). Analysing these properties still faces serious computational complexity. Reduction of model size was proposed as a mean to cope with this issue. Furthermore, to enhance the capacity of Boolean networks to produce relevant predictions, formal methods have been developed to systematically identify control strategies enforcing desired behaviours. In their paper, E. Tonello and L. Paulevé [4] assess the most popular reduction that consists in eliminating a model component. Considering three typical update schemes (synchronous, asynchronous and general asynchronous updates), they thoroughly study the effects of the reduction on attractors, minimal trap spaces (minimal subspaces from which the model dynamics cannot leave), and on phenotype controls (interventions which guarantee that the dynamics ends in a phenotype defined by specific component values). Because they embody potential behaviours of the biological process under study, these are all properties of great interest for a modeller. The authors show that eliminating a component can significantly affect some dynamical properties and may turn a control strategy ineffective. The different update schemes, targets of phenotype control and control strategies are carefully handled with useful supporting examples. Overall, E. Tonello and L. Paulevé’s contribution underlines the need for caution when defining a regulatory network and characterises the consequences on critical model properties when discarding a component [4]. References [1] Motoyosi Sugita (1963) Functional analysis of chemical systems in vivo using a logical circuit equivalent. II. The idea of a molecular automation. Journal of Theoretical Biology, 4, 179–92. https://doi.org/10.1016/0022-5193(63)90027-4 [2] Stuart Kauffman (1969) Metabolic stability and epigenesis in randomly constructed genetic nets. Journal of Theoretical Biology, 22, 437–67. https://doi.org/10.1016/0022-5193(69)90015-0 [3] René Thomas (1973) Boolean formalization of genetic control circuits. Journal of Theoretical Biology, 42, 563–85. https://doi.org/10.1016/0022-5193(73)90247-6 [4] Elisa Tonello, Loïc Paulevé (2024) Phenotype control and elimination of variables in Boolean networks. arXiv, ver.2 peer-reviewed and recommended by PCI Math Comp Biol https://arxiv.org/abs/2406.02304 | Phenotype control and elimination of variables in Boolean networks | Elisa Tonello, Loïc Paulevé | <p>We investigate how elimination of variables can affect the asymptotic dynamics and phenotype control of Boolean networks. In particular, we look at the impact on minimal trap spaces, and identify a structural condition that guarantees their pre... |  | Dynamical systems, Systems biology | Claudine Chaouiya | 2024-06-05 10:12:39 | View |
FOLLOW US
MANAGING BOARD
Wolfram Liebermeister
Celine Scornavacca (Representative)
Donate Weghorn










