
Systematic investigation of software tools and design of a tailored pipeline for paleoproteomics research
Benchmarking the identification of a single degraded protein to explore optimal search strategies for ancient proteins
Abstract
Recommendation: posted 17 October 2024, validated 21 October 2024
Assis, R. (2024) Systematic investigation of software tools and design of a tailored pipeline for paleoproteomics research. Peer Community in Mathematical and Computational Biology, 100309. https://doi.org/10.24072/pci.mcb.100309
Recommendation
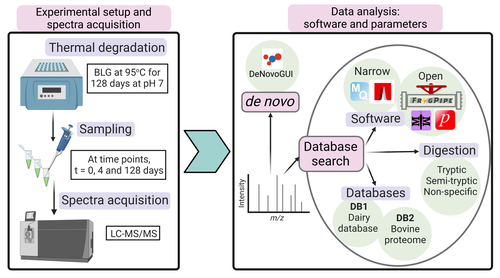
Paleoproteomics is a rapidly growing field with numerous challenges, many of which are due to the highly fragmented, modified, and degraded nature of ancient proteins. Though there are established standards for analysis, it is unclear how different software tools affect the identification and quantification of peptides, proteins, and post-translational modifications. To address this knowledge gap, Rodriguez Palomo et al. design a controlled system by experimentally degrading and purifying bovine beta-lactoglobulin, and then systematically compare the performance of many commonly used tools in its analysis.
They present comprehensive investigations of false discovery rates, open and narrow searches, de novo sequencing coverage bias and accuracy, and peptide chemical properties and bias. In each investigation, they explore wide ranges of appropriate tools and parameters, providing guidelines and recommendations for best practices. Based on their findings, Rodriguez Palomo et al. develop a proposed pipeline that is tailored for the analysis of ancient proteins. This pipeline is an important contribution to paleoproteomics and is likely to be of great value to the research community, as it is designed to enhance power, accuracy, and consistency in studies of ancient proteins.
References
Ismael Rodriguez-Palomo, Bharath Nair, Yun Chiang, Joannes Dekker, Benjamin Dartigues, Meaghan Mackie, Miranda Evans, Ruairidh Macleod, Jesper V. Olsen, Matthew J. Collins (2023) Benchmarking the identification of a single degraded protein to explore optimal search strategies for ancient proteins. bioRxiv, ver.3 peer-reviewed and recommended by PCI Math Comp Biol https://doi.org/10.1101/2023.12.15.571577
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
This work has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie grant agreement No 956410
Reviewed by anonymous reviewer 1, 07 Oct 2024
I thank Palomo et al. for their thoughtful responses to my reviews. The physicochemical section is excellent. I'm happy that you included the isoelectric point and other properties. I think it really strengthens the observed results and will be helpful for similar future preservational studies on other proteins.
https://doi.org/10.24072/pci.mcb.100309.rev21Reviewed by Shevan Wilkin , 17 Oct 2024
, 17 Oct 2024
The authors have adjusted all of the comments I requested. The paper reads well and is extremely thorough. I would recommend the manuscript at this point.
The only “issue” I have is that the proposed pipeline includes several searches using multiple platforms for each sample, which is exponentially increases the time and work to complete any study. Hopefully some researchers incorporate this pipeline into new work so we can see the how much these additional steps add to the reliability of ancient protein studies.
https://doi.org/10.24072/pci.mcb.100309.rev22Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2023.12.15.571577
Version of the preprint: 2
Author's Reply, 20 Sep 2024
Decision by Raquel Assis, posted 23 Jul 2024, validated 24 Jul 2024
The submitted manuscript is an important contribution to paleoproteomics. However, the reviewers have identified several issues with the clarity of the manuscript text and figures. I recommend that the authors carefully address these on a point-by-point basis, particularly as this manuscript should serve as a guide for best practices in paleoproteomics.
Reviewed by anonymous reviewer 1, 03 May 2024
Reviewed by Shevan Wilkin , 22 Jul 2024
, 22 Jul 2024
Please see my attached document with comments. Please feel free to contact me with any questions. Apologies if anything isn't clear - I just returned from holiday am down with COIVD...
Shevan
Download the review https://doi.org/10.24072/pci.mcb.100309.rev12









